News
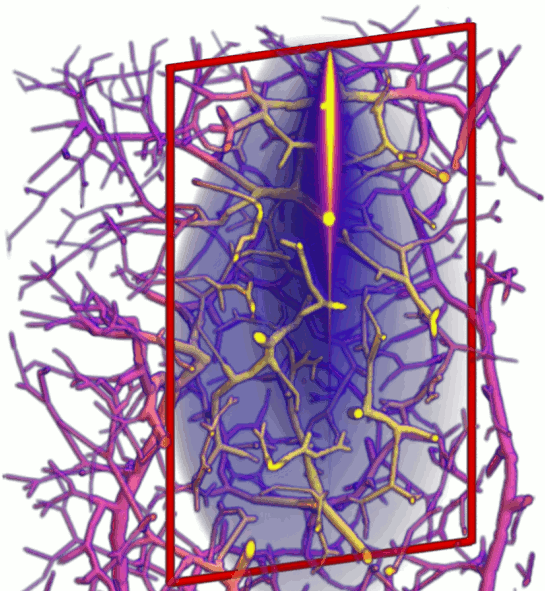
New paper preprint - implicit MMC for complex tissues
October 11, 2020
Checkout our newly submitted paper on a highly scalable and memory efficient MMC approach to model very complex tissue structures. The implicit MMC (iMMC) code was developed by PhD student Yaoshen Yuan, and will be available in our next MMC release.
Light transport modeling in highly complex tissues using implicit mesh-based Monte Carlo algorithm
Also check out this cool animation: https://twitter.com/FangQ/status/1315417111708069888
Split-voxel MC (SVMC) paper is published
October 9, 2020
Congrats to my student @Shijie_Yan for the new paper on Split-Voxel MC (SVMC). It is nearly as good as MMC and as efficient as MCX.Full citation information:
Shijie Yan and Qianqian Fang, "Hybrid mesh and voxel based Monte Carlo algorithm for accurate and efficient photon transport modeling in complex bio-tissues," Biomed. Opt. Express 11, 6262-6270 (2020)
Code will be released shortly.
MCX and MMC v2020 have arrived
September 6, 2020
MCX/MMC/MCXCL v2020 was announced. They have packed numerous commits accumulated over the past 16 months, including the first MCX-CL release that matches MCX in features, the first GPU-based MMC, win installer, and saving #openjdata outputs.
Get your copy from http://mcx.space/wiki/?Get here].

MCX R01 was successfully renewed!
August 1, 2020
With the kind support from many of our users, the MCX grant has been successfully renewed! Over the past 5 years, the MCX project has grown tremendously. We have more than quadrupled the total number of registered users (500 to 2400) and the number of citations (250 to 1100). Many of the new methods and features have been developed, published, and added to our open-source toolkit; mcx/mmc are now available in Fedora and Winget, and will soon become available in Debian/Ubuntu. Our software has found use in a wide range of applications and was increasingly tested and trusted by many. We feel truly honored to contribute our small part to many of your endeavors, and would be very much eager to share our new development with you down the path. In the next chapter of the MCX project, we have planed many exciting new milestones, many of which are already under heavy development. Stay tuned, more will come!
See our road-map for the next 4 years.
Try our new all-in-one Windows installer for MCX/MMC
July 5, 2020
We created a Windows all-in-one installer for our MCX/MMC users to automate the installation and settings on Windows. It contains both binaries and MATLAB toolboxes for all of our software components.
Nightly build installer is at http://mcx.space/nightly/win64/
Give it a try!
See screenshots in this tweet: https://twitter.com/FangQ/status/1279935567095087106
Photon-sharing paper is published on Optics Letters
May 12, 2020
Congratulations to our PhD student Shijie Yan for publishing a new paper on Optics Letters. In this method, one can simultaneously simulate many pattered light sources using our MCX/MMC simulators. This feature is now supported in our open-source software.
Shijie Yan, Ruoyang Yao, Xavier Intes, and Qianqian Fang, "Accelerating Monte Carlo modeling of structured-light-based diffuse optical imaging via “photon sharing”," Opt. Lett. 45, 2842-2845 (2020)
Download PDF from our preprint on biorxiv.
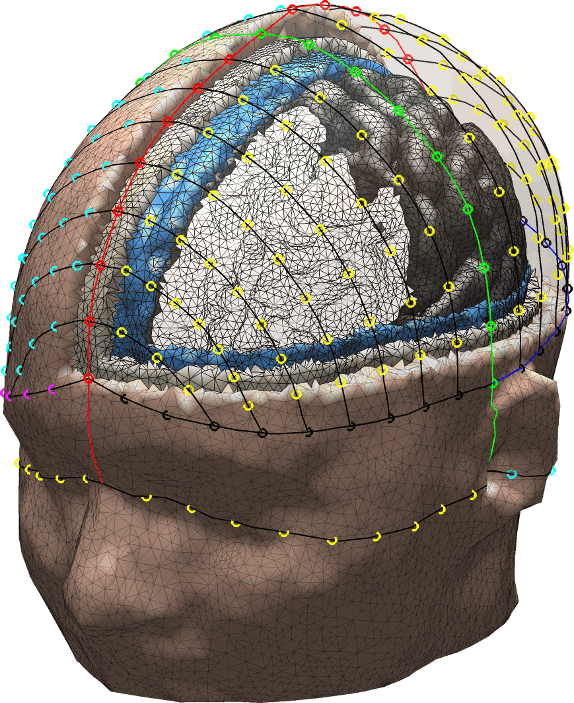
Brain2Mesh paper is published in Neurophotonics
February 5, 2020
Brain2Mesh is a MATLAB/Octave based 3D mesh generation toolbox dedicated to the creation of high-quality multi-layered brain mesh models. The details of this toolbox is described in this newly published paper on Neurophotonics:
Anh Phong Tran†, Shijie Yan†, Qianqian Fang*, (2020) "Improving model-based fNIRS analysis using mesh-based anatomical and light-transport models," Neurophotonics, 7(1), 015008
Brain2mesh is open-source! Download it from http://mcx.space/brain2mesh/
New paper on photobiomodulation dosage over lifespan is published in Neurophotonics
February 5, 2020
Our PhD student Yaoshen Yuan's latest paper on photobiomodulation dosage over a wide span of age groups, ranging from 5 to 89 years old, is now published in Neurophotonics:
Yuan Y, Cassano P, Fang Q*, (2020) “Transcranial Photobiomodulation with Near-Infrared Light from Childhood to Elderliness: Simulation of Dosimetry",Neurophotonics, 7(1), 015009
GPU MMC paper is published on JBO
November 20, 2019
Our OpenCL based GPU-accelerated mesh-based Monte Carlo (mmcl) is now published on JBO:
Qianqian Fang* and Shijie Yan, “Graphics processing unit-accelerated mesh-based Monte Carlo photon transport simulations,” J. of Biomedical Optics, 24(11), 115002 (2019)
The software is also available, see our mailing list announcement, and speedup over different devices.
JData Specification Draft 1 is complete
May 13 2019
JData is a general-purpose data interchange format aimed for portability, readability and simplicity. It utilizes the JavaScript Object Notation (JSON) [RFC 4627] and Universal Binary JSON (UBJSON) specifications to store complex hierarchical data in both text and binary formats. In this specification, we define a list of JSON-compatible constructs to store a wide range of data structures, including scalars, arrays, structures, tables, hashes, linked lists, trees and graphs, and support optional data grouping and metadata for each data element. The generated data files are compatible with JSON/UBJSON specifications and can be readily processed by most existing parsers. Advanced features such as array compression, data linking and anchoring are supported to greatly enhance portability and scalability of the generated data files.
The Draft-1 of the specification is now available at https://github.com/fangq/jdata
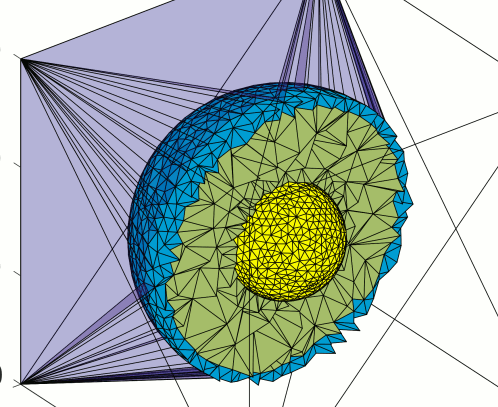
A new letter paper on dual-mesh Monte Carlo algorithm is published on JBO
February 20, 2019
The paper focuses on the speed improvement of mesh-based MC (DMMC) photo transport simulator by using a coarsely tessellated tetrahedral mesh for ray-tracing computation and an independent voxelated grid for output data storage. DMMC is able to improve the speed by 1.3 × to 2.9 × for various scattering settings. This paper is now available online.
Shijie Yan, Anh Phong Tran, Qianqian Fang, "Dual-grid mesh-based Monte Carlo algorithm for efficient photon transport simulations in complex three-dimensional media," J. Biomed. Opt. 24(2) 020503 (20 February 2019) https://doi.org/10.1117/1.JBO.24.2.020503
DMMC has been integrated into current version of MMC.
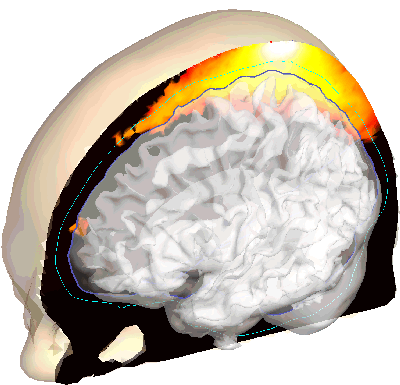
A new paper on photobiomodulation (PMB) dosimetry is in press on Neurophotonics
Janunary 7, 2019
In collaboration with Prof. Paolo Cassano from MGH, we performed a Monte Carlo simulation based study on the illumination positions (transcranial and intranasal) and 3D dosimetry for photobiomodulation (low-light therapy) using our in-house simulators to better guide the design of PMB treatment procedures and devices. This paper is in press on Neurophotonics.
Tran AP+, Cassano P+, Katnani H, Bleier BS, Hamblin MR, Yuan Y, Fang Q*, (2019) “Selective photobiomodulation for emotion regulation: model-based dosimetry study,” Neurophotonics 6(1) 015004, PMCID: PMC6366475
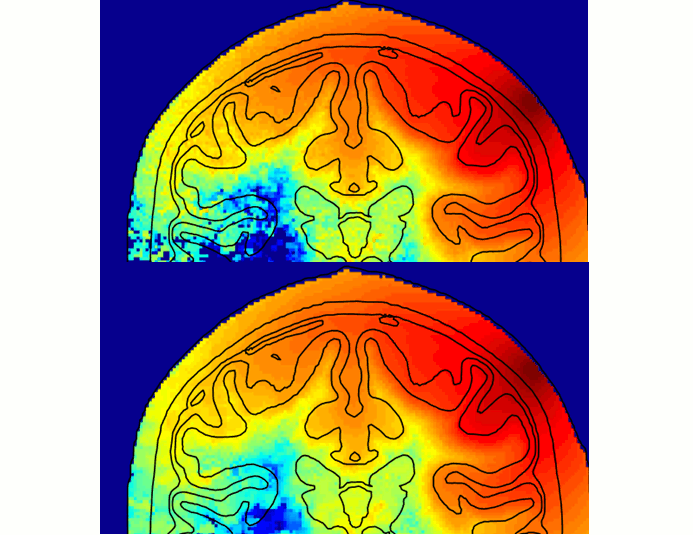
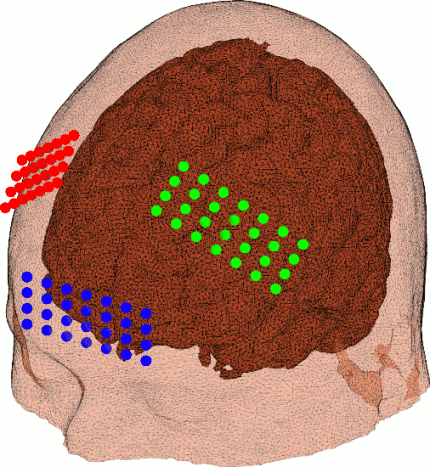
New paper on Monte Carlo simulation denoising
November 30, 2018
Our paper on denoising Monte Carlo simulation data is now available online at the JBO website in the coming issue. PDF. Full citation information:
Yaoshen Yuan, Leiming Yu, Zafer Doğan, Qianqian Fang*, "Graphics processing units-accelerated adaptive non-local means filter for denoising three-dimensional Monte Carlo photon transport simulations," J. of Biomedical Optics, 23(12), 121618 (2018).
Open source code in Github.
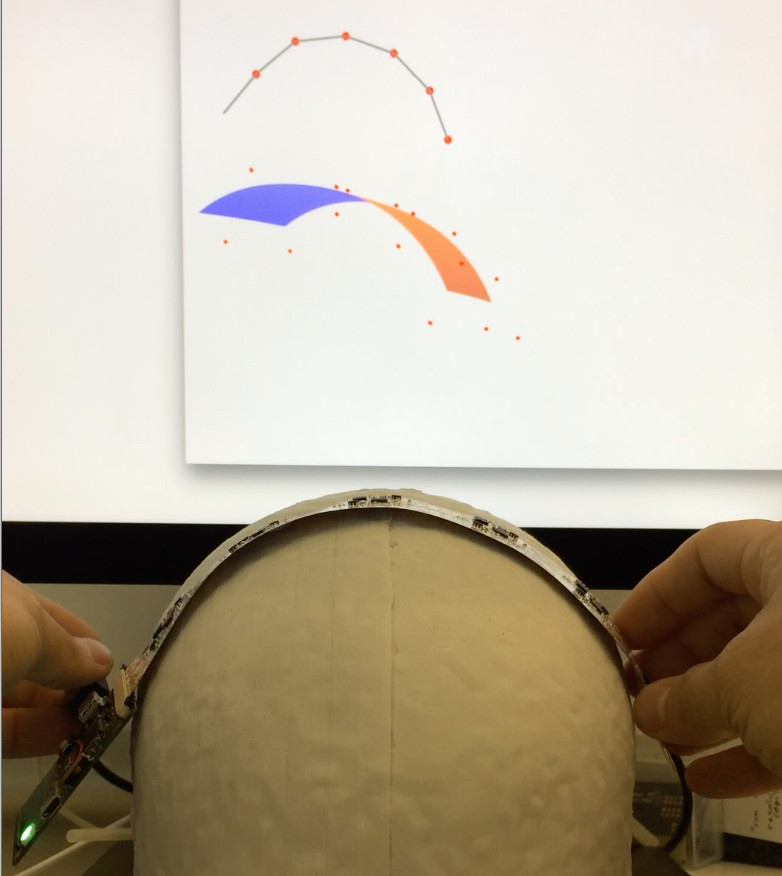
fNIRS Conference 2018
October 7th, 2018
Fanglab presented their work towards a fiberless, modular, and wearable full-head fNIRS system with built-in 3D self-calibrating orientation sensor network at the 2018 fNIRS Conference held in Tokyo, Japan.Find the abstract in our Publications page

Fanglab was awarded a new NIH R01 grant for next-gen fNIRS
September 21, 2018
The project aims to advance optical brain imaging to the next generation by developing an ultra-portable, modular, and fiberless optical brain imaging platform. The proposed imaging system can be used to monitor human brain activities in natural environments.

MCX 1.0 and MMC 1.0 have arrived!
August 24, 2018
After nearly 10 years of continuous development, it is our great pleasure to announce that MCX 1.0 (v2018) has finally arrived! In the meantime, MMC has also arrived at its v1.0 milestone, after its initially publication in 2010. Moreover, we also proudly announce the first stable release of MCX-OpenCL (or MCXCL) - a "clone" of MCX that can run on nearly all CPUs and GPUs across many vendors.
Demo of MCX at Redhat Summit 2018
May 07, 2018
Researchers at the Boston Children's Hospital (Jason Sutin, Ivy Lin, Ellen Grant, and Julia Tatz) had ported MCX to the cloud using the Redhat Kubernetes and containers, and Prof. Ellen Grant will give a demo of the software during the Redhat Developer Conference this Wed.

FangLab students presented 9 posters at OSA Biophotonics Congress
April 3, 2018
Our group presented 9 posters at the OSA Biophotonics Congress: check out our published conference papers (PDF available)
- Yuan Y, Yu L, and Fang Q*, “Denoising in Monte Carlo Photon Transport Simulation Using GPU-accelerated Adaptive Non-Local Mean Filter,” OSA BIOMED 2018, Paper#JTh3A.41, FL
- Yu L, Nina-Paravecino F, Kaeli D, Fang Q*, “Fast Monte Carlo Photon Transport Simulations for Heterogeneous Computing Systems,” OSA BIOMED 2018, Paper#2912790, FL
- Vanegas M, Zimmerman B, Sahin S, Carp S, Fang Q*, “A Mobile Phone Based Reflectance Pulse Oximeter,” OSA BIOMED 2018, Paper#JTu3A.64, FL
- Vanegas M, Carp S, Fang Q*, “Mobile Phone Camera Based Near-Infrared Spectroscopy Measurements,” OSA BIOMED 2018, Paper#JTu3A.64, FL
- Yan S, Tran AP, Fang Q*, “A Dual-Mesh Monte Carlo Algorithm Using a Coarse Tetrahedral Mesh and Voxel Output”, OSA BIOMED 2018, Paper# JTh3A.17, FL
- Tran AP, Fang Q*, “Generating High Quality Tetrahedral Meshes of the Human Head and Applications in fNIRS,” OSA BIOMED 2018, Paper#JTu3A.52, FL
- Ruoyang Y, Intes X, Fang Q*, “Efficient Jacobian Construction for Hyperspectral Wide-field Diffuse Optical Tomography via Replay Monte Carlo,” OSA BIOMED 2018, Paper#JTu3A.30, FL
- Xu E, Fang Q*, “Optical Tomography Reconstruction to Recover Small Oriented Inclusions from Optically-Thick Diffusive Media”, OSA BIOMED 2018, Paper#JTu3A.67, FL
- Sahin S†, Sun X†, Vanegas M, Tran AP, Fang Q*, “Developing an Anatomically Accurate Multi-layered Optical Brain Phantom for fNIRS Studies”, OSA BIOMED 2018, Paper#JW3A.70, FL

MCX Workshop 2018 at OSA Biophotonics Congress
April 2, 2018
This year, the MCX workshop was held during the first day of the OSA Biophotnics Conference in Hollywood Florida. We implemented a lot of feedback from the 2017 workshop in Boston to provide what we hope were helpful tips, insight, and knowledge on how to best take advantage of our in-house developed high-performance Monte Carlo photon transport simulation platforms. Relive the experience!
Fang Lab won a TIER1 seed grant!
March 16, 2018
Dr. Fang and https://web.northeastern.edu/amylu/ Dr. Amy Lu] received a https://www.northeastern.edu/resdev/resources/internal-grants/tier-1-seed-grantproof-of-concept-program/ "TIER1" seed grant] from Northeastern University to develop the next generation wearable fNIRS imaging system.
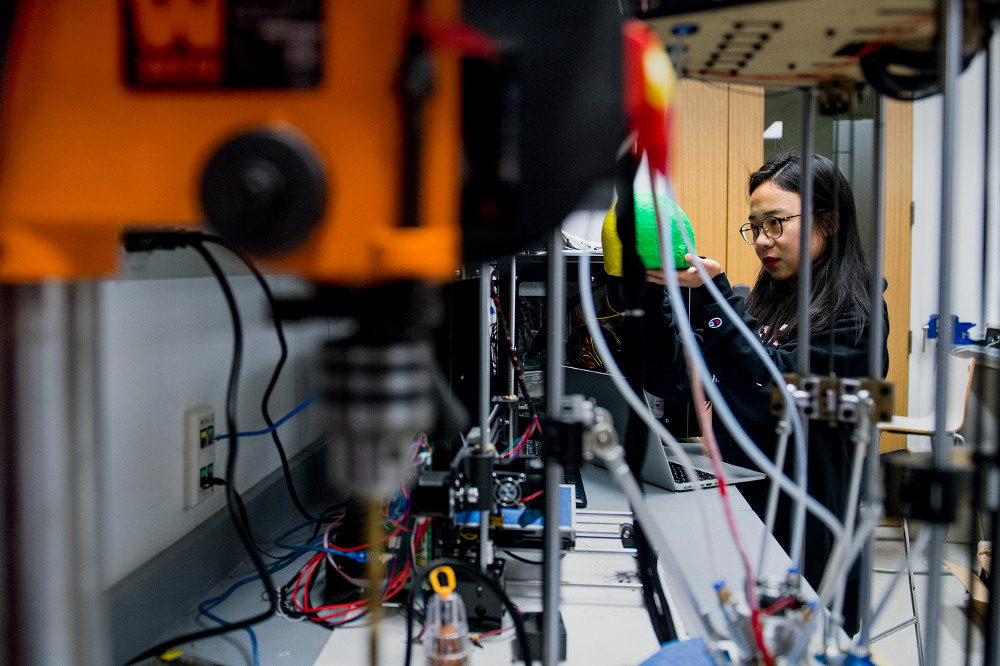
Xin made the Northeastern News!
March 16, 2018
Our graduate student Xin Sun was featured in the Northeastern News working on our anatomically accurate multi-layered full-head optical phantom (together with our Bioengineering undergraduate student Sule Sahin).

Morris Named 2018 MIT Impact Fellow
January 15, 2018
Morris Vanegas was just named a 2018 MIT Impact Fellow! The program is tailored towards career development for post-doctoral and advanced pre-doctoral trainees. The group and one-on-one mentoring helps fellows articulate the impact of their research to a brach audience and help identify actionable opportunities to strengthen their work focus. See details of the Impact Program here.

Morris Named 2018 Dent Scholar
November 9, 2017
Morris Vanegas was just named a 2018 Dent The Future Scholar! This award comes with a complimentary pass to Dent 2018, a flagship conference for a community of entrepreneurs, executives and creatives who are driven to "put a dent in the universe." Dent looks for scholars who work at the intersection of art, science, and impact. Morris was chosen for his work at co-director of the MIT Open Mind::Open Art Gallery in January 2017. See details of Dent 2018 here.
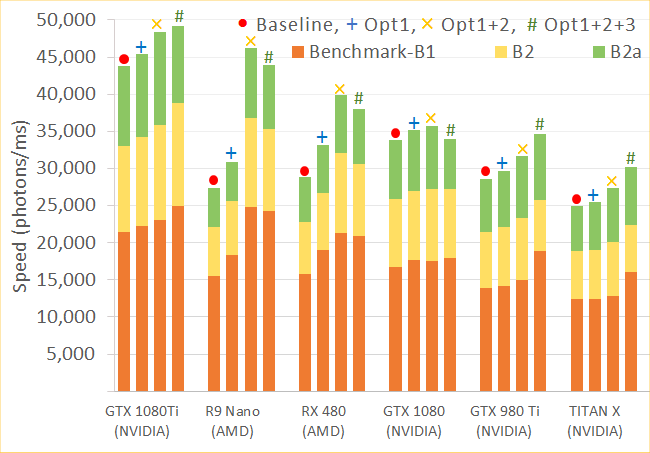
New Paper Published
January 26, 2018
Our paper is now published online at the JBO website in the coming issue. [ PDF ]. Full citation information: Leiming Yu, Fanny Nina-Paravecino, David Kaeli, Qianqian Fang, "Scalable and massively parallel Monte Carlo photon transport simulations for heterogeneous computing platforms," J. Biomed. Opt. 23(1), 010504 (2018).
November 4, 2017
A manuscript entitled "Scalable and massively parallel Monte Carlo photon transport simulations for heterogeneous computing platforms" was submitted to a journal for consideration of publication. This paper summarizes our over 7 years continuous development of MCX-CL - the OpenCL version of MCX designed for expanding our GPU accelerated MC simulation to NVIDIA/AMD/Intel processors using the OpenCL framework.Download the manuscript preprint PDF or software.
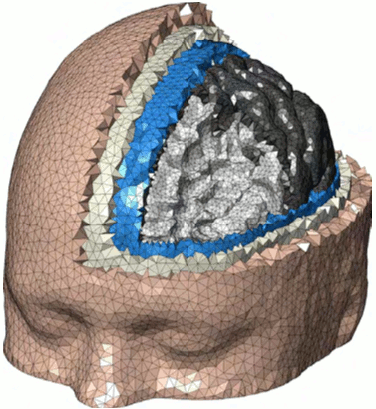
New Paper Submission
August 29, 2017
A manuscript entitled, "Fast and high-quality tetrahedral mesh generation from neuroanatomical scans" was submitted to a journal. The pre-print of the manuscipt is available at http://arxiv.org/abs/1708.08954.
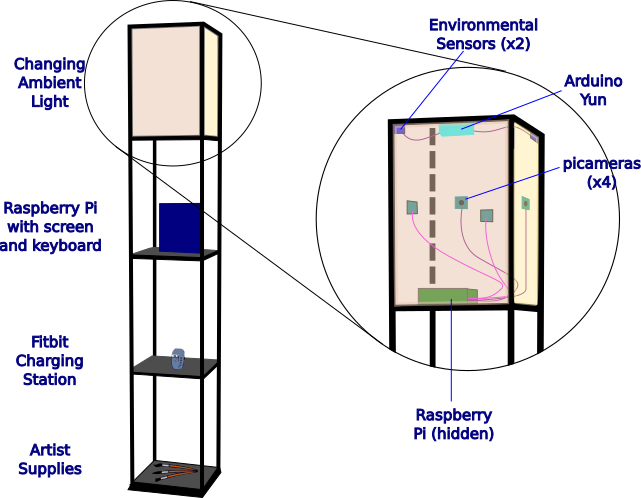
IDEA Venture Accelerator Prototype Funding Winner!
August 22, 2017
Morris Vanegas was recently awarded funds to complete his prototype of a portable Internet of Things (IoT) suite of sensors to capture temporal artist physiological and environmental data by Northeastern's IDEA Venture Accelerator. The stand-alone 3 shelf stand and light also uses OpenCV to non-invasively capture an artist's work. The images and data are then presented in an interactive GUI.

MCX Workshop 2017 at ISEC
August 8-9, 2017
Thanks to those that attended our 1st two-day workshop on Monte Carlo eXtreme for biomedical optics research at ISEC on August 8th and 9th. We hope we were able to provide our software users with tips, insight, and knowledge on how to best take advantage of our in-house developed high-performance Monte Carlo photon transport simulation platforms. Many thanks to our speakers and COTI student presenters, as well as to the attendees for their active participation and discussions. We plan on making this an annual event, so feel free to reach out if you want to attend in the future. See the images below to relive the experience!For more details, including tutorials from the event, click here.
Build your own GPU rack!
In support of our MCX 2017 workshop, Dr. Fang and Morris built a rack that houses 12 GPUS, 3 power supplies, and is made of 80-20s extrusion bars. Users will be able to run monte-carlo simulations by remoting into these machines. A quick test of 5x 1080Ti GPUs with a single power supply was able to simulate 311955.21 photons per ms, making it our fastest setup to date!We've also documented the process of building the GPU rack below:
Looking for a postdoc position?
As our lab continues to expand, we are looking for highly qualified candidates for two postdocs in the near-term for NIH-funded research projects. If you, or someone you know, is a great fit and would like to join our team located at ISEC, reach out to us!Optics Engineering We are also looking for a postdoc with a background in optical instrumentation, medical device design, and mobile-app development. A strong electrical engineering background would be a plus. Apply here!
Computational Biophotonics This position is perfect for someone with a strong background in computational algorithm development, GPU computing, and computational physics. Proficiency in C/matlab software development is a necessity. Apply here!
![[Home]](upload/lab_full_logo.png)